Parameter selection with CroptimizR
Samuel Buis
2026-02-10
Source:vignettes/Parameter_selection.Rmd
Parameter_selection.RmdIntroduction
This vignette illustrates a general workflow for parameter
estimation and selection in crop models using CroptimizR and
CroPlotR.
The workflow presented here implements the parameter selection algorithm
described in Wallach
et al. (2023), which provides a detailed methodology for stepwise
parameter estimation.
This parameter estimation and selection algorithm proceeds through a sequence of successive steps. Its core idea is to distinguish between major parameters, which are systematically estimated in order to reduce bias, and candidate parameters, whose estimation is optional.
Major parameters are chosen for their strong and consistent influence on model outputs across all environments. A good candidate for this category typically affects simulated values in all environments.
Candidate parameters are those expected to explain part of the remaining variability between environments, but for which it is not always clear whether estimating them will improve model performance.
In the algorithm, major parameters are estimated first using an Ordinary Least Square (OLS) criterion. Candidate parameters are then added one by one to the set of estimated parameters and retained only if they lead to an improvement in an information criterion value (e.g., Akaike or Bayesian Information Criterion). Candidate parameters that are not selected are kept fixed at their default values.
Candidate parameters should be ordered, as far as possible, according to decreasing expected importance. While there is no strict limit on the number of candidate parameters, increasing this number directly increases the computational cost of the procedure.
Using an information criterion helps the algorithm limit overfitting, because it is calculated as the sum of a term measuring the fit of simulated values to observations and a penalty that increases with the number of estimated parameters.
Study Case
The crop model input data used in this example comes from a maize crop experiment (see description in Wallach et al., 2011). To illustrate the calibration procedure, julian days of two phenological stages were simulated from this dataset on 8 different environments (called situations in CroptimizR language) and then used as observations, after adding some random errors.
The STICS crop model is used in this example. Initialization steps
required by the use of this model (definition of the
model_options argument of the estim_param
function) are hidden in this example for sake of brevity. They are
detailed in the vignette Parameter
estimation with CroptimizR: a simple case using the STICS crop
Model.
Note however that CroptimizR can be applied to any crop model, provided that a suitable wrapper is available. Guidelines and examples for implementing wrappers are provided in the Designing a model wrapper vignette.
Wrappers already exist for several models. For more details, see the Get started page or contact the authors.
Plotting the observations
The observations are provided here in the obs_list
object, in the format required by CroptimizR, i.e. as a named list of
data frames (or tibble), each element being named after the
corresponding situation (see the Get
started vignette of CroptimizR for details).
In this example, the variables corresponding to the two observed
stages considered are called iamfs and ilaxs.
They correspond to the julian days (from the beginning of the sowing
year) of “end juvenile” and “maximum LAI” stages as defined in the STICS
crop model.
# Show the observations for the first two situations
obs_list[1:2]## $`bo96iN+`
## Date iamfs ilaxs
## 1 1996-10-15 168 230
##
## $bou00t1
## Date iamfs ilaxs
## 1 2000-11-01 181 221The 8 situations are called here bo96iN+,
bou00t1, bou00t3, bou99t1,
bou99t3, lu96iN+, lu96iN6 and
lu97iN+.
Let’s plot the values of these observations:
obs_df <- dplyr::bind_rows(obs_list) %>% tidyr::pivot_longer(
cols = c("iamfs", "ilaxs"), names_to = "stage"
)
ggplot(obs_df, aes(x = stage, y = value)) +
geom_boxplot()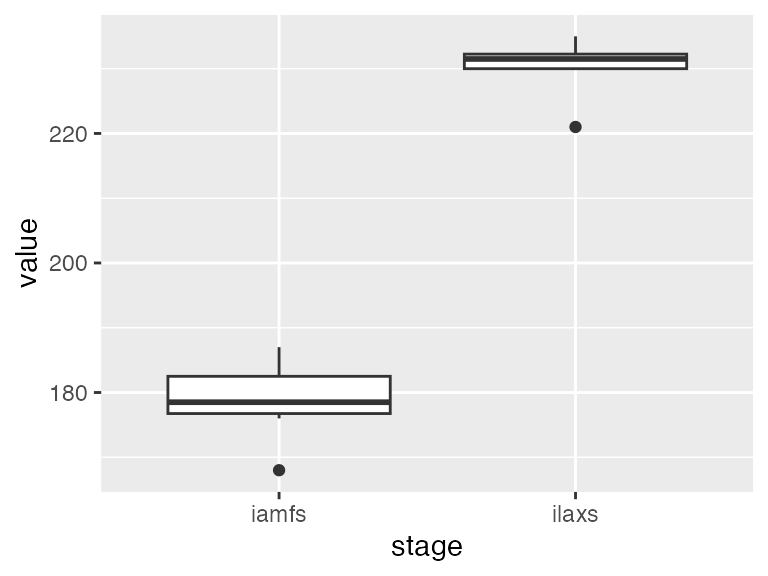
The “end juvenile” stage occurred around day 180 after sowing, while the “maximum LAI” stage occurred around day 230 after sowing.
Setting information on the parameters to estimate
The whole list of parameters to estimate and their lower and upper
bounds (noted resp. lb and ub) must be
provided in the param_info argument of the
estim_param function.
In this example, we will consider 5 parameters of the STICS crop
model: stlevamf, stamflax, tdmin,
stressdev and tdmax.
param_info <- list(
lb = c(stlevamf = 100, stamflax = 300, tdmin = 4, stressdev = 0, tdmax = 25),
ub = c(stlevamf = 500, stamflax = 800, tdmin = 8, stressdev = 1, tdmax = 32)
)Lower or upper bounds can be set to -Inf or
Inf, respectively. In such cases, initial values must be
provided for the corresponding parameters (see ?estim_param
for further details).
Candidate parameters must be specified through the
candidate_param argument of the estim_param
function. This vector must be ordered from those thought to be the most
influential on the considered variable, to the least influential. This
ordering is important, as it directly determines the sequence in which
candidate parameters are considered and tested during the stepwise
parameter selection procedure.
In this example, 3 parameters were considered as candidates.
candidate_param <- c("tdmin", "stressdev", "tdmax")Parameters that are included in param_info but not
listed in candidate_param are treated as major parameters
and are therefore systematically estimated at all steps of the
procedure. In this example, stlevamf and
stamflax belong to this category and are estimated
regardless of the selected candidate parameters.
Choosing the default parameters values
Default values for the non-estimated parameters and for the candidate
parameters can be defined in the forced_param_values
argument of the estim_param function.
forced_param_values <- c(tdmin = 5.0, stressdev = 0.2, tdmax = 30.0)These values will be used at each step for which these parameters are not estimated.
Setting optimization options
During the first step of the algorithm, all major parameters are estimated simultaneously, starting from randomly selected initial values. For subsequent steps, the initial values of parameters that have already been estimated are fixed at their previously estimated values, and only the initial value of the newly added candidate parameter is randomly drawn. As a consequence, Wallach et al. (2023) recommend performing a larger number of optimization repetitions for the first step (typically 10), and fewer repetitions for the subsequent steps (typically 5). This strategy is implemented as follows:
Running the optimization
A single call to the estim_param function is sufficient
to run the complete parameter estimation and selection procedure.
Compared with the simple example presented in https://SticsRPacks.github.io/CroptimizR/articles/Parameter_estimation_simple_case.html
, it is important here to explicitly define the
crit_function argument. The stepwise parameter selection
procedure illustrated in this vignette is currently implemented for an
Ordinary Least Squares objective function, provided by
crit_ols, which is not the default criterion used by
estim_param.
Note also the use of the forced_param_values and
candidate_param arguments, which respectively control the
values of non-estimated parameters and the sequential inclusion of
candidate parameters during the selection process.
By default, estim_param uses the corrected Akaike
Information Criterion (AICc) for parameter selection. In this example,
the Bayesian Information Criterion (BIC) is used instead, as specified
through the info_crit_func argument.
res <- estim_param(
obs_list = obs_list,
model_function = model_wrapper,
model_options = model_options,
crit_function = crit_ols,
optim_options = optim_options,
param_info = param_info,
forced_param_values = forced_param_values,
candidate_param = candidate_param,
info_crit_func = list(CroptimizR::BIC),
out_dir = data_dir
)At the end of the execution, some information such as the selected step number, the list of selected parameters, their estimated values and some stats about the execution time are printed in the R console:
...
# -- Summary of the Parameter Selection procedure results
#
# Selected step: 2
# Selected parameters: stlevamf,stamflax,tdmin
# Estimated value for stlevamf : 321.18
# Estimated value for stamflax : 589.73
# Estimated value for tdmin : 7.96
#
# A table summarizing the results obtained at the different steps is stored in C:/Users/sbuis/AppData/Local/Temp/RtmpwRzZkP/data-master/study_case_1/V9.0/param_selection_steps.csv
# Graphs and detailed results obtained for the different steps can be found in C:/Users/sbuis/AppData/Local/Temp/RtmpwRzZkP/data-master/study_case_1/V9.0/param_select_step# folders.
#
# Average time for the model to simulate all required situations: 4.1 sec elapsed
# Total number of criterion evaluation: 806
# Total time of model simulations: 3293 sec elapsed
# Total time of parameter estimation process: 3314 sec elapsed
# ----------------------In this case, the object returned by the estim_param
function contains the results corresponding to the selected step, as
well as a data frame named param_selection_steps that
provides detailed information for all steps of the parameter selection
procedure. For each minimization step,
param_selection_steps reports the set of estimated
parameters, their initial and final values, and the associated values of
the sum of squares and information criteria. The selected step is
identified in the last column of this data frame. For convenience, the
same information is also written to a CSV file
(param_selection_steps.csv), saved either in the current
working directory or in the directory specified by the
out_dir argument of estim_param.
| Estimated.parameters | Initial.parameter.values | Final.values | Initial.Sum.of.squared.errors | Final.Sum.of.squared.errors | BIC | Selected.step |
|---|---|---|---|---|---|---|
| stlevamf, stamflax | 356.22, 522.43 | 404.58, 739.23 | 2960 | 147 | 41.03 | |
| stlevamf, stamflax, tdmin | 404.58, 739.23, 7.67 | 321.18, 589.73, 7.96 | 4242 | 91 | 36.13 | X |
| stlevamf , stamflax , tdmin , stressdev | 321.18, 589.73, 7.96, 1.00 | 321.18, 589.73, 7.96, 1.00 | 91 | 91 | 38.90 | |
| stlevamf, stamflax, tdmin , tdmax | 321.18, 589.73, 7.96, 30.20 | 321.18, 589.73, 7.96, 31.07 | 91 | 91 | 38.90 |
In this example, the selected parameter set consists of
stlevamf, stamflax, and tdmin,
corresponding to the lowest value of the BIC among all tested steps.
The complete object returned by the estim_param function is also
saved to disk in the file optim_results.Rdata.
Standard result files and diagnostic plots produced by
estim_param for each step of the procedure are stored in
the param_select_step# folders.
Generating diagnostics using CroPlotR
To generate diagnostic plots, the model wrapper is first run using
the estimated values of the selected parameters, while all remaining
parameters are set to their default values. These values are passed to
the model through the param_values argument.
param_values <- c(res$final_values, res$forced_param_values)
sim_after_optim <- model_wrapper(
param_values = param_values,
model_options = model_options,
var = c("iamfs", "ilaxs")
)Once the simulations have been performed, simulated and observed values can be compared graphically using the plotting functions provided by CroPlotR.
In particular, scatter plots of simulated versus observed values can be produced as follows:
plot(sim_after_optim$sim_list, obs = obs_list, type = "scatter")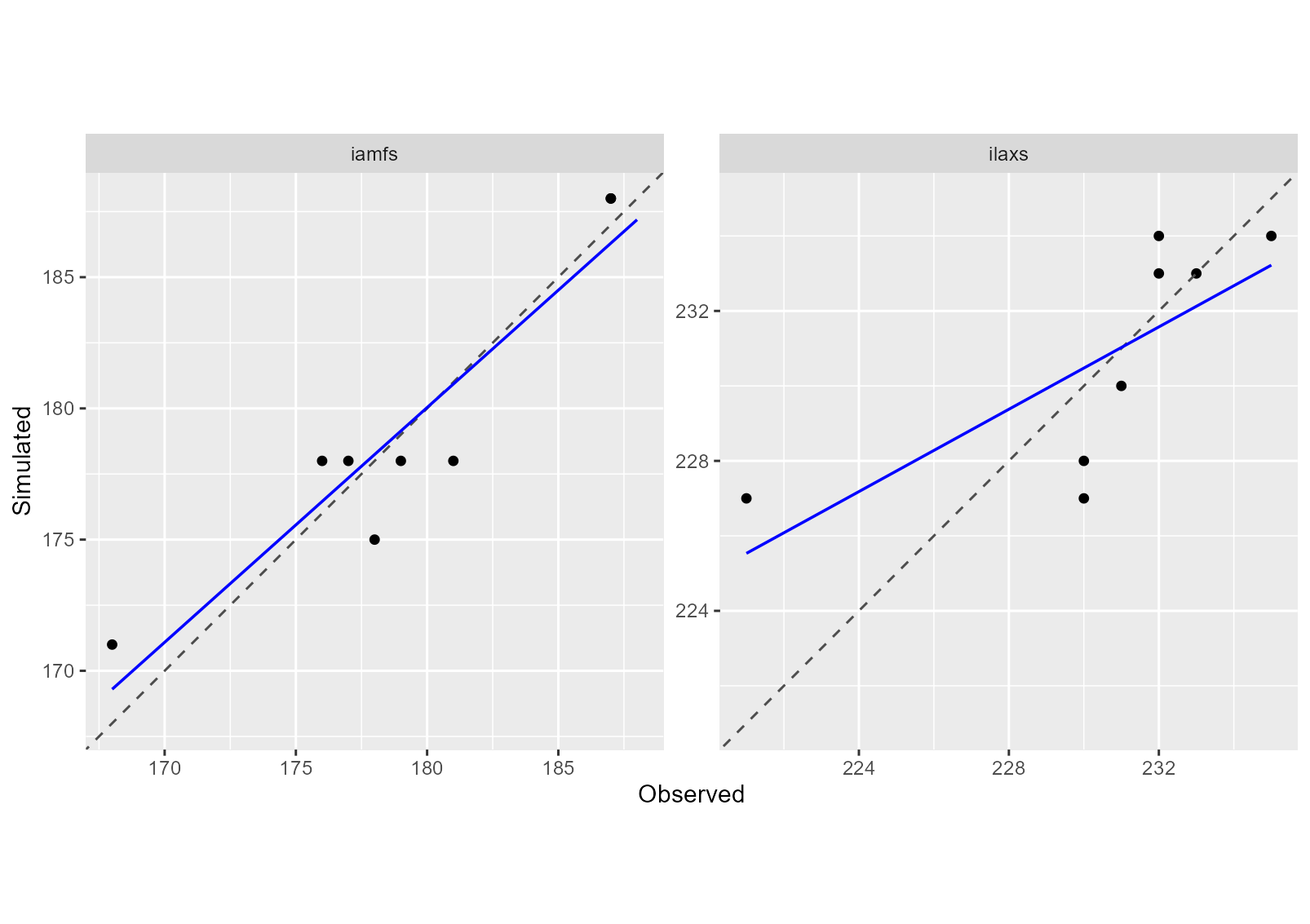
In addition to graphical diagnostics, quantitative performance
metrics can be computed for each variable. In this example, the Mean
Squared Error (MSE) and its decomposition are calculated using the
summary function from CroPlotR.
## # A tibble: 2 × 7
## group situation variable MSE Bias2 SDSD LCS
## <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
## 1 Version_1 all_situations iamfs 4.38 0.0156 0.0637 4.92
## 2 Version_1 all_situations ilaxs 7 0.0625 1.14 6.78